Copper Circuitry
CueR, a member of the MerR protein family, responds to zeptomolar concentrations of free copper and is the most sensitive of the copper sensors in enterobacteria. Members of the MerR metalloregulatory protein family exert tight control over transcription and act at a single site within the prokaryotic core promoter both as repressors and as ligand-inducible activators of transcription. The O’Halloran group determined the structure of CueR/DNA with and without bound metal to resolve the paradox of how CueR can both repress and activate transcription. Crystal structures of the CueR-DNA complexes reveal how the repressor and activator torque and bend DNA in this genetic switching event to alter the distance between the -35 and -10 promoter elements to prevent or promote binding of RNA polymerase (Figure 2). It is this new overall topology that activates transcription, rather than re-phasing of the -10 and -35 promoter elements.
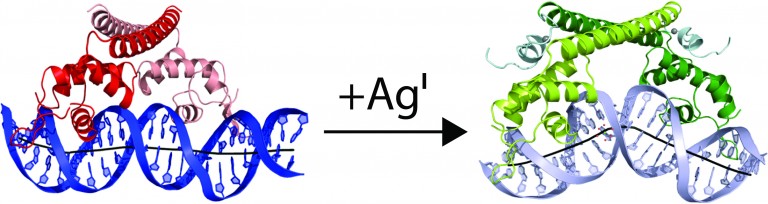
Figure 2: CueR alters the topology of DNA. In the absence of metal, CueR dimer (red) binds to a 33 bp fragment of the copA promoter to repress transcription. Addition of metal switches CueR to the activator state (green). In the activator state, CueR changes the topology of copA to promote binding of RNA polymerase. (Philips et al., 2015).
Selected Publications:
- Philips SJ, Canalizo-Hernandez M, Yildirim I, Schatz GC, Mondragón A, O’Halloran TV. “Allosteric transcriptional regulation via changes in the overall topology of the core promoter”. Science. 2015 ;349(6250):877-81.
- Outten, F.W., Outten, C., Hale, J., and O’Halloran, T. “Transcriptional Activation of an E. coli Copper Efflux Regulon by the Chromosomal MerR Homologue, CueR.” J. Biological Chemistry, 2000;275 (40):31024-31029.
-
Ansari AZ, Bradner JE, O’Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374(6520):371-5.
-
Ansari, A.Z., Chael, M.L., O’Halloran, T.V. “Allosteric Underwinding of DNA is a Critical Step in Positive Control of Transcription by MerR.” Nature. 1992;355:87-89.